Make high-performance bioinformatics accessible
Rime Bioinformatics develops services to accelerate medical, agronomic and veterinary research to develop alternatives to antibiotics.
In addition to the time-consuming in vivo and in vitro phases, these research projects require in silico phases which can hamper research teams´ progress. Tools for analyzing DNA or RNA sequencing data are indeed tricky to use for biologists and computer scientists: they require advanced skills in computer science and biology that are difficult to master for a single person. The significant computing power required to run these abundant and continuously updated softwares is an additional obstacle to access precision analyses.
However, bioinformatics analyzes play a key role in the development of new safe human, veterinary or agricultural drugs. Regulatory bodies such as the EMA and the FDA thus require extensive analyzes before putting new remedies to market. If these studies are not of satisfactory quality, market access may be slowed down or blocked. For a more efficient research aimed at patient safety and environment preservation, accessible and high-performance bioinformatics analyzes are needed.
Fighting antibiotic resistance with phage therapy
Antibiotics are our main weapon to fight bacterial infections, in human and veterinary health, as well as in agronomy. Their benefits go beyond saving patients: the protection they provide to farms safeguards the incomes of farmers and the food security of the communities that depend on them.
However, bacterial resistance to antibiotics is pushing the latter towards decline. Without a solution, the UN predicts 10 million deaths each year in 2050 and a cost for humanity between 2015 and 2050 estimated at 100,000 billion USD.
Bacteriophages (or phages) are bacteria-killing viruses, discovered at the beginning of the 20th century. Phage therapy can be used to control bacteria in many areas: human health, food safety, agriculture, breeding, cosmetics. They are an effective and more environmentally friendly alternative to antibiotics.
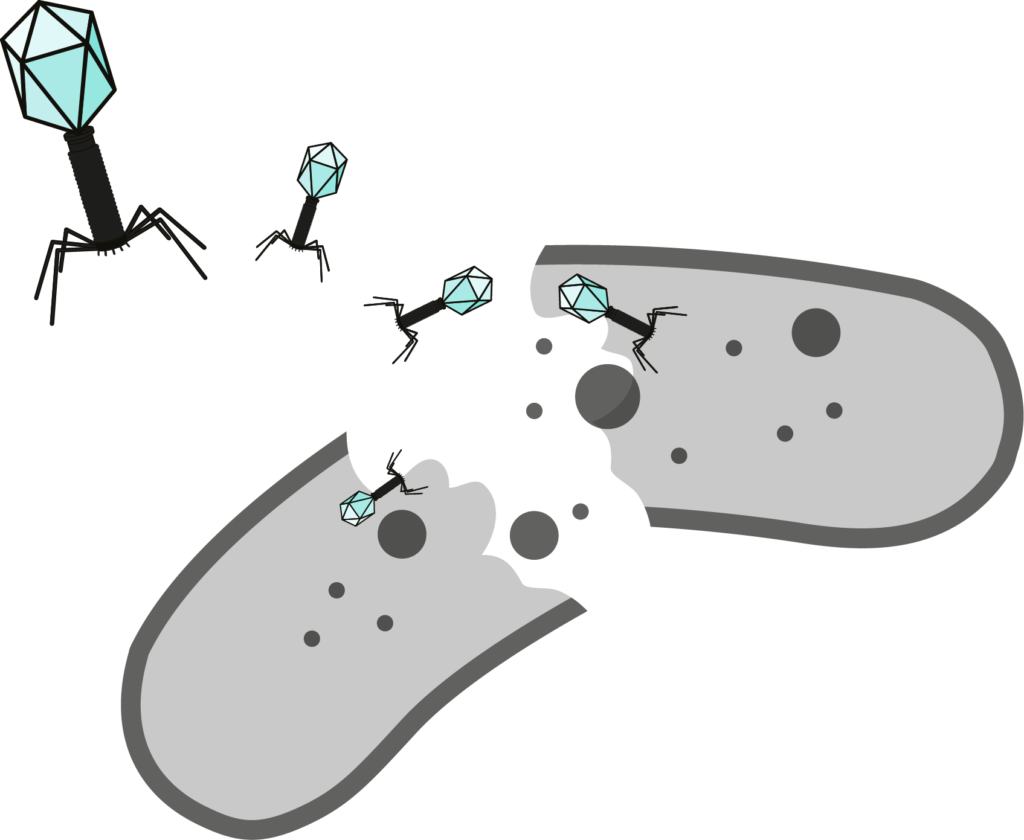
Bioinformatics, a key step to phage therapy implementation
Bioinformatics is the science of analysing data yielded by life science research. Applied to phage therapy research, it helps to understand phage nucleic acids (RNA or DNA) and to predict phage’s biological behaviors. Before using a phage as anti-bacterial treatment, it is essential to study it in vivo, in vitro and in silico. Such analysis is necessary to assess a phages’ safety for patients or the environment. However, currently published phage genomes leave an important part of unknown. For example, less than 24% of functions of genes from phages that infect Vibrio spp. bacteria are known. To promote a faster and safer development of these alternatives to antibiotics, we developed a range of analysis methods that allow a precision study of phage genomes.
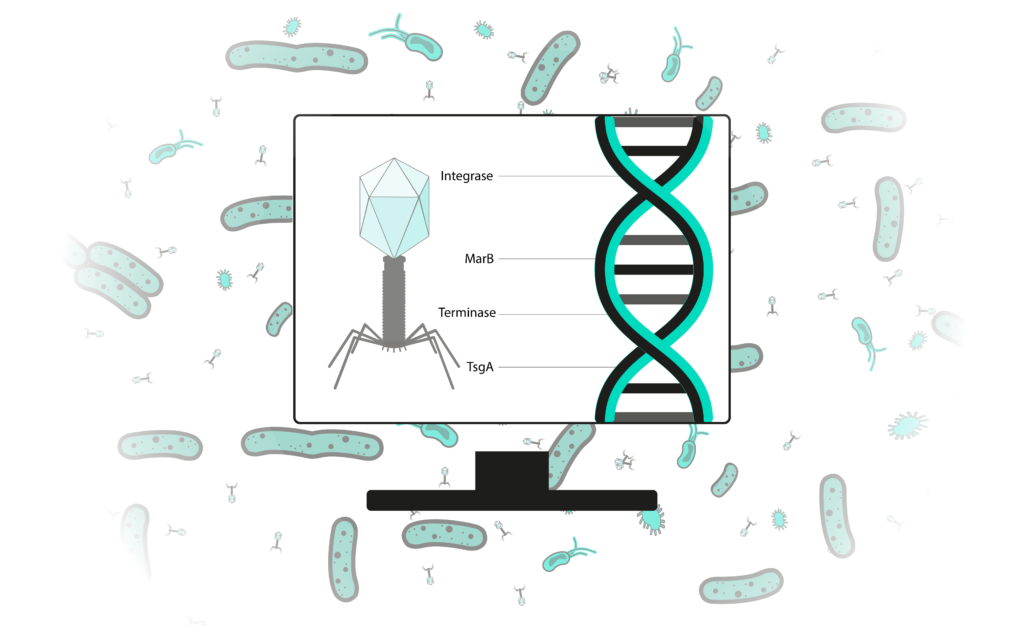
Bringing performance to bioinformatics services
Bioinformatics tools are typically difficult to install, use complex user interfaces and can require powerful computers to run. These softwares are the output of a very active research field: they evolve quickly and are endlessly replaced by new ones. This specificity hampers biologists’ access to quality analysis.
Our mission is to make high performance bioinformatics accessible to biologists. To fulfill that purpose, we continually follow and benchmark the latest advances of the field and develop our own tools. These help us produce precise results. The later are delivered in clear reports, aimed at the biologist we work with.

Our partners


















