Phage genome annotation
Get the most out of your sequence data for your R&D and your Proof of Concept
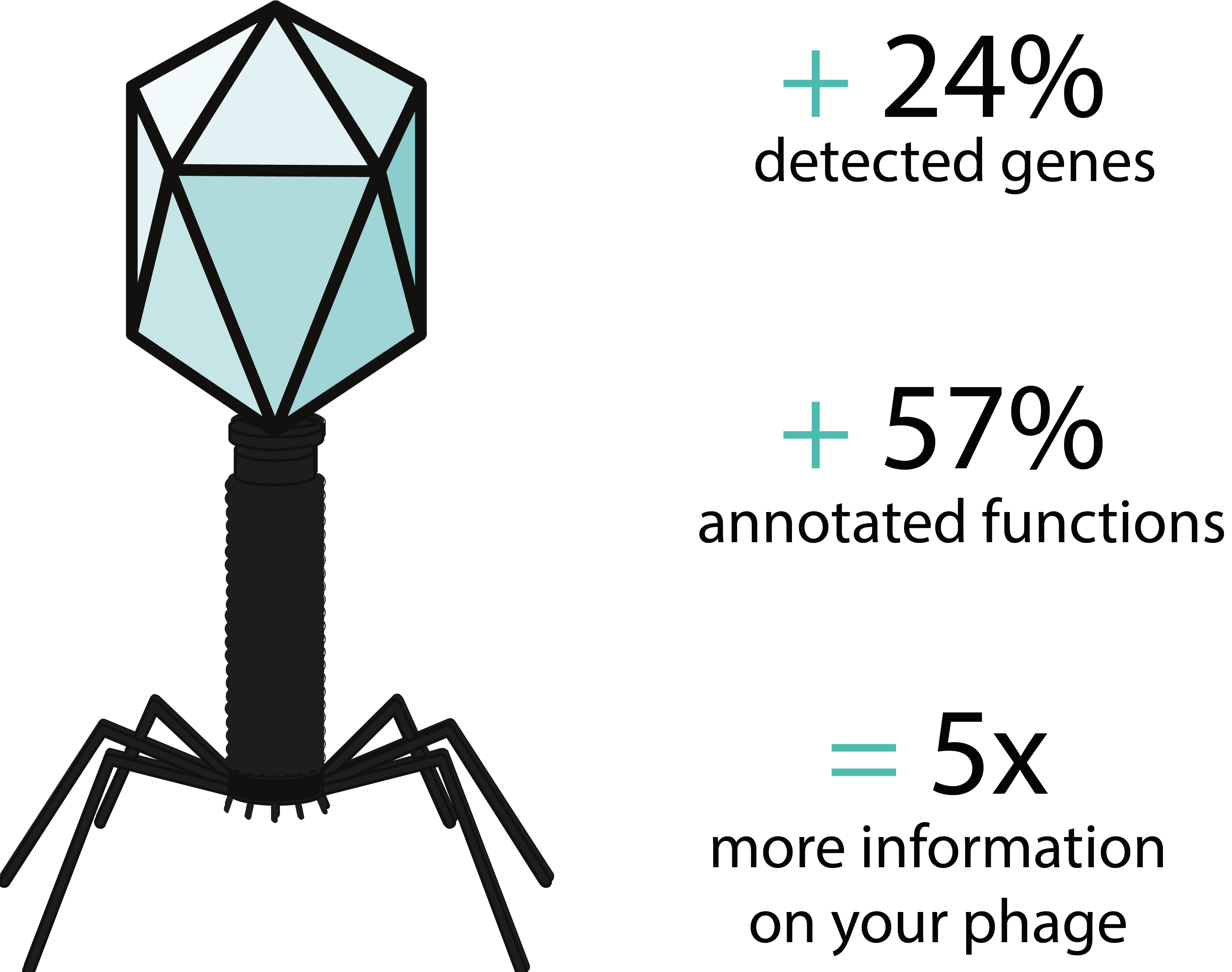
* Results are based on the comparison of the performance of our pipeline against a standard annotation method for the study of a published phage genome. The genome of Vibrio vulnificus phage VVP001, published by Kim et al (2021) was used.
At every step of the analysis, the robustness of our annotations arises from the comparison of results yielded by multiple methods.
Our pipeline’s high sensitivity for gene function detection is enabled by the numerous and comprehensive databases we use.
Finally, expert manual annotation provides finer results and brings a global understanding of the genome.
* Results are based on the comparison of the performance of our pipeline against a standard annotation method for the study of a published phage genome. The genome of Vibrio vulnificus phage VVP001, published by Kim et al (2021) was used.

At every step of the analysis, the robustness of our annotations arises from the comparison of results yielded by multiple methods.

Using multiple databases allows a high gene function detection sensitivity.
Manual annotation provides a finer study on regions of interest and brings a global understanding of the genome.
Why should you use our services ?
European and Northern American regulators demand extensive evidence dossiers. Backing your proof of concept up with a comprehensive genome analysis, such as ours, is key for clearance.
Our expertise comes from research lab experience with phages, bacteria, and their genomes. Staying at the edge of scientific research is of prime importance to us. This is why we participate in ongoing projects with university research groups.
Concepts and methods used in bioinformatics can be cryptic. As we want you to fully take advantage of your genome data, our mission is to perform high quality analysis and provide easily understandable results.
